Get Aozan 3.1.1
 Download Aozan Application
Download Aozan Application
Welcome to Aozan
Aozan has been developed in order to automatically handle raw data transfer, demultiplexing and quality control of a sequencing run once an Illumina sequencer run has been completed. This software is involved in the primary data analysis. Aozan produces compressed FASTQ files and a quality report from an Illumina sequencer output in order to evaluate each sequencing parameter. Aozan can work on most computer infrastructures and only requires a samplesheet for each run.
Aozan greatly helps to improve the efficiency in the run data management and to keep tracks of the run statistics through automatic mails and HTML report pages.
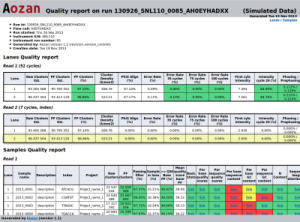
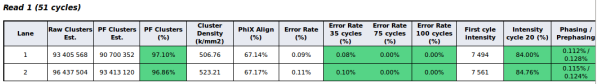
Quality control report example
Supported devices
Aozan can handle the output of many Illumina sequencer models, however not all the models of Illumina sequencers has been tested with Aozan. As Illumina sequencer outputs are very similar, Aozan may work with most of the Illumina sequencers. You can contact us to tell us if your sequencer model is compatible with Aozan.
| RTA version | Model | Support status |
|---|---|---|
| <1.18.64 | HiSeq 1000, HiSeq 1500, HiSeq 2000, HiSeq 2500, MiSeq | Supported and tested (HiSeq 1000 and HiSeq 1500) |
| 1.18.64 and 2.1.x - 2.6.x | NextSeq 500, HiSeq X, HiSeq 1000, HiSeq 1500, HiSeq 2000, HiSeq 2500 | Supported and tested (NextSeq 500 and HiSeq 1500) |
| 2.7.x | HiSeq 3000, HiSeq 4000, HiSeq X, MiniSeq | Supported and tested (HiSeq 3000) |
| 3.7.x or above | NextSeq 1000 and NextSeq 2000 | Supported and tested (NextSeq 2000) |
What's new in Aozan 3?
Support for NextSeq 1000/2000
Aozan now support NextSeq 1000/2000 sequencers and RTA 3.x.
Support for BCL Convert
Aozan now support BCL Convert in addition of bcl2fastq.
Major code rewriting
Aozan 3 is now a full Java application. All the existing Python code has been rewitten in Java. With this major code rewrite, Aozan will be able to support other sequencing technologies.
What's new in Aozan 2.2?
MultiQC support
Aozan can now generate a MultiQC report for each run project.
To activate the MultiQC report creation, just enable the qc.test.project.multiqc.report.enable parameter in your configuration file.
What's new in Aozan 2.0?
Bcl2fastq 2.x support
Aozan can now handle bcl2fastq 2.x and its new samplesheet file format. Aozan do not support anymore bcl2fastq 1.x as bcl2fastq 2.x can handle all Illumina sequencer output since HiSeq 1000/2000. Aozan has been tested with bcl2fastq 2.16, 2.17 and 2.18.
FastQC version 0.11.5
FastQC version 0.11.5 is now bundled in Aozan.
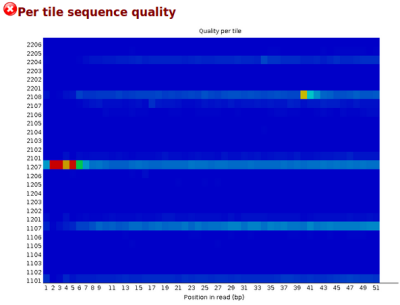
Example of a report of the new "Per tile sequence quality" module of FastQC:
Enhancement to FastQC
The control quality steps contains an additional FastQC module named "Bad tiles" that search tiles with BMS (Bottom Middle Swath) issues with HiSeq flowcell v3.
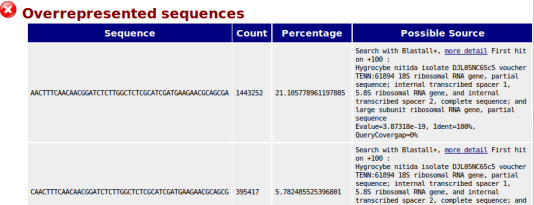
Optionally, Aozan adds a sub-step in FastQC to enhance the Overrepresented sequences module. For sequences with "no hit", a blastn can be launch to estimate the source. Blastn results with 100% identity and 0% gap will be included in report.
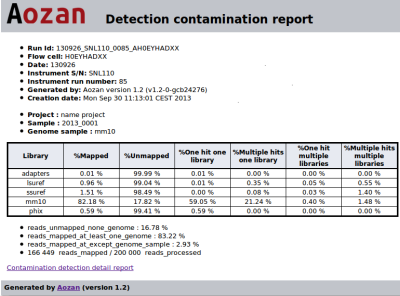
Contamination detection with FastQ-Screen
Aozan now include a fast Java FastQ Screen implementation. This module maps reads samples on a list of reference genomes for assessing sample contamination and the ratio of the expected genome in the sample. It creates a report file with values for each genome.
FastQC and contamination dectection on undetermined FASTQ files
Two parameters have been added in the Aozan configuration file to run FastQC and contamination detection on the undetermined FASTQ files. For the contamination detection, all the available genomes related to the run will be used.
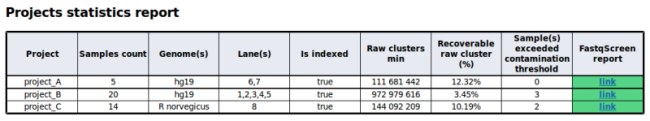
New projects tests in quality control report
The quality control report has been enhanced with a new project section. This new section gather sample data according their project in the run. You will find more details in the project tests section of the documenatation.
An example of table built with projects tests.
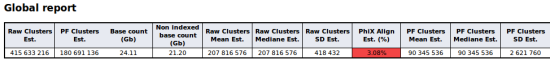
New globals tests in quality control report
The quality control report has been enhanced with a new global section. This new section contains global information about the run. You will find more details in the global tests section of the documentation.
An example of table built with global tests:
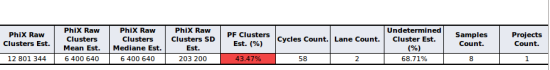
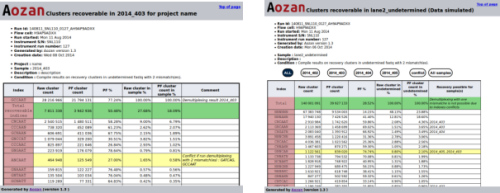
Analysis of the indices of reads in the undetermined FASTQ files
Aozan now provide a QC module that analyzes indices of reads in the undetermined FASTQ files. For each indices, this module suggest the sample(s) and the number of reads that can be recovered using a demultiplexing step with one more mismatch that the number of mismatches has been used. See for more information about this feature the sample tests section of the documentation.
The reports generated by this module show for each sample, the list of indices and read counts that can be recovered.
New built-in step to automatically recompress all your fastq files.
Aozan has now a new built-in step which recompresses FASTQ files (compressed or not with gzip) into bzip2 files. bzip2 files are more compressed than gzip files and thus take less disk space.
Warning: The recompression step has been removed in Aozan 3.
Define denied runs and prioritized runs
In the aozan.var.path directory, you can specify [step].deny files (e.g. qc.deny, demux.deny...) that contains the list of runs to not process by the step.
You can also create in this directory a runs.priority file that contains the list of the runs to process before the others runs.
Furthermore, when a sequencer run fail, it will be automatically added to the hiseq.deny file.
Aozan can now fully be executed using a Docker image.
Aozan can now fully be executed using a Docker image. Thus you don't need anymore to install Aozan dependancies on your computer before using it.
Bcl2fastq samplesheet location
If a bcl2fastq samplesheet is not provided in the samplesheet directory, Aozan will use if exist the Samplesheet.csv file located at the root of raw output run directory.
Number of mismatch for demultiplexing
The bcl2fastq samplesheets can now contain a setting with the number of allowed mismatch for demultiplexing.
Aozan can now manage new Illumina HiSeq and NextSeq sequencers
Aozan can manage several sequencers, and now handles the latest versions of Illumina sequencers (HiSeq 3/4000 and NextSeq 500).
Demultiplexing using docker
Aozan can launch bcl2fastq 2.x in Docker container. Bcl2fastq 2 Docker images are available on our dockerhub.
New include directive in configuration file
The Aozan configuration file has been improved with a new "include" directive.
With this enhancement, it will be easier to use Aozan in a multi-servers context using the same core configuration file by all Aozan instances.
Reading Illumina InterOp binary files
Run data are now retrieved by parsing Illumina InterOp Binary Metric files as Illumina XML run files are now deprecated.
New Samplesheet Validator v2
A new samplesheet validator is available, it supports now samplesheet for bcl2fastq 2.x.
Continuous synchronization of HiSeq data
Aozan can now perform continuous synchronizations of working runs to avoid a big synchronizations at the end of the runs.
Availability
Aozan is distributed under the General Public License and CeCill.
Funding
This work was supported by the Infrastructures en Biologie Santé et Agronomie (IBiSA) and France Génomique.